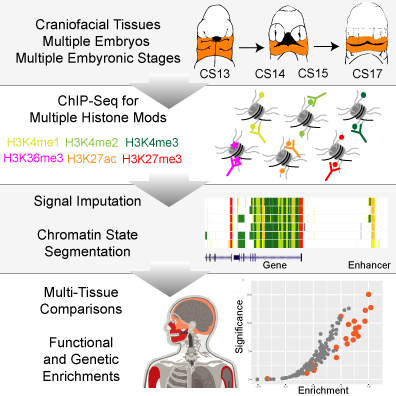
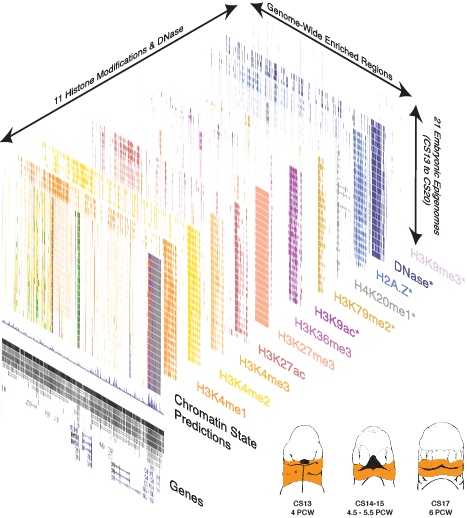
Epigenomic Project now published in Cell Reports! https://doi.org/10.1016/j.celrep.2018.03.129
Epigenomic Project Original preprint on biorxiv.org: http://biorxiv.org/content/early/2017/05/10/135368
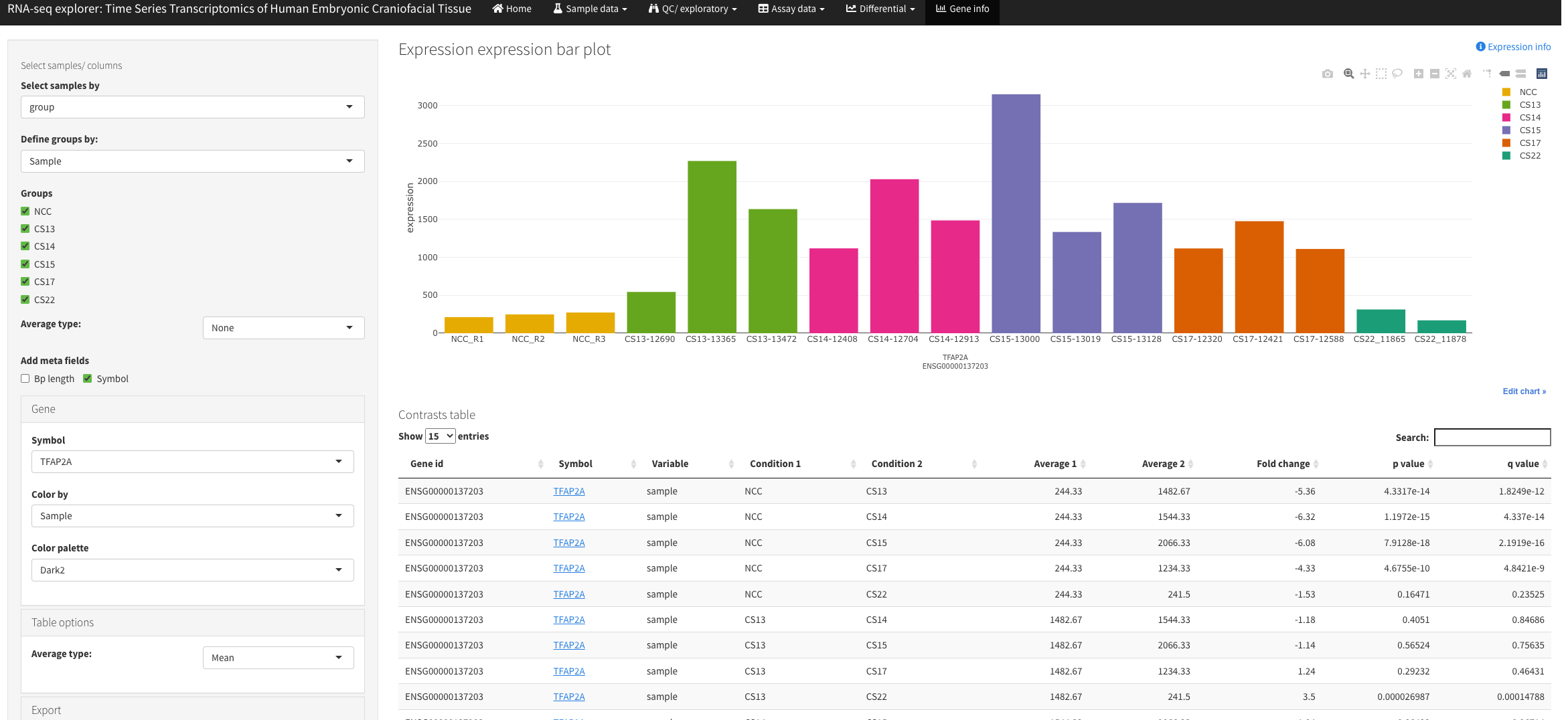
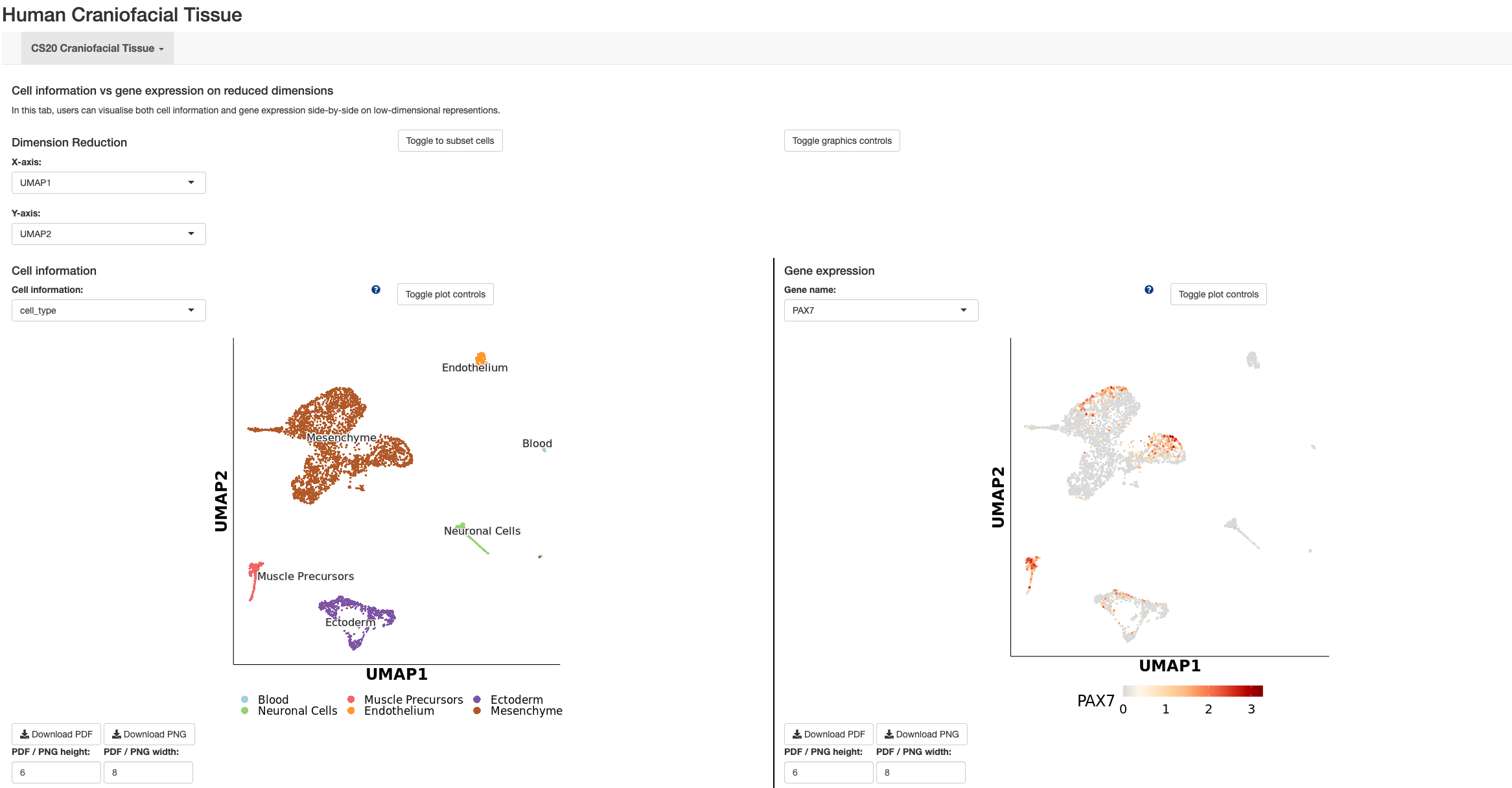
Transcriptomic Project preprint on biorxiv.org: https://doi.org/10.1101/2022.02.28.482338
Updates:
02-28-2022 All chromatin state annotations have been converted to hg38 coordinates. RNA-Seq data from Yankee et al. 2022 (forthcoming) are now available. See links below for RNA-Seq data exploration apps.
Video Tutorial for UCSC Track Hubs
Individual Links
Load our data into the UCSC genome browser using track hub functionality (all data in hg19):
- Human Craniofacial ChIP-Seq and Chromatin State Segmentations: hub file
Jump to your favorite gene in a new browser window (Please email for more gene-specific links):
- 8q24 Vista Enhancers
- ABCA4 Vista Enhancers
- ALX3 Vista Enhancers
- ALX4 Vista Enhancers
- ARHGAP29 Vista Enhancers
- BMP4 Vista Enhancers
- DLX1 Vista Enhancers
- FGFR2 Vista Enhancers
- FOXE1 Vista Enhancers
- IRF6 Vista Enhancers
- MAFB Vista Enhancers
- MSX1 Vista Enhancers
- MSX2 Vista Enhancers
- NOG Vista Enhancers
- NTN1 Vista Enhancers
- PAX7 Vista Enhancers
- PRDM16 Vista Enhancers
- PTCH1 Vista Enhancers
- TWIST1 Vista Enhancers
- TWIST2 Vista Enhancers
- TXNDC16 Vista Enhancers
- VAX1 Vista Enhancers
- Orthologous 4C-Seq Interaction data at the HoxA cluster here
- 4C-Seq Interaction data at the HoxA cluster in mouse here
All Figures and Tables are available on FigShare.
Imputed Signal Tracks and Chromatin State Segmentations are available on GEO: GSE97752
Mouse 4C-Seq data are available on GEO: GSE98251
Epilogos
Open our data in HiGlass